BiGGr Models
BiGGr Models is a knowledgebase of genome-scale metabolic network reconstructions. It is an evolution of the BiGG Models repository (legacy BiGG ).
Overview
BiGG Models integrates a plethora of published and unpublished genome-scale metabolic networks into a single database with a set of standardized identifiers, called BiGG IDs. Genes in the BiGG models are mapped to NCBI genome annotations whenever possible, metabolites are anchored to reference compounds (typically ChEBI or InChI ), and reactions are linked to RHEA reactions, if available.
Source Code
Database
BiGGr presents metabolic models as standardized database entries. To help understand the database representations and standardizations, the different types of database entities are explained below. Yellow boxes describe major differences or improvements compared to legacy BiGG. Thumbnail images are shown which link to examples of the described entity or behaviour.
Models & Collections

A Model represents an entity that combines metabolites, reactions, and genes into a form that can be used to model aspects of an organism or tissue or group thereof. In BiGGr, Models are members of a Collection. Collections group models that were constructed in the same research, pipeline or manuscript together. In cases where a single model was constructed in a specific research, the models still belong to a distinct collection, but this collection is often omitted (e.g. in the new collections page). The BiGG IDs of models should still be globablly unique, i.e. a model can always be accessed by its BiGG ID alone, without the necessity of knowing the collection BiGG ID.
Metabolites
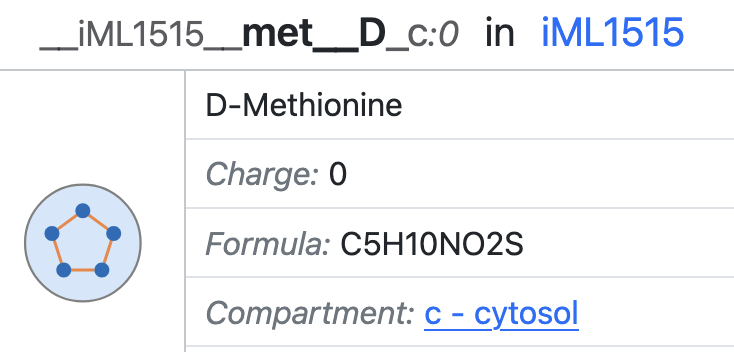
A Metabolite (or Component) represents a chemical compound present in a model. A component is linked to one Reference Compound (or multiple in the case of tautomers). A Reference Compound represents an (external) definition of the metabolite. In most cases this is a ChEBI entity. However, in cases where no ChEBI is available, a compound can be anchored to a InChI string. Components that represent the same compound in different protonation states are grouped together under the entity Universal Component. Each Universal Component has a default Component, represents the Component the database will default to when no explicit charge is given.

Universal Component have BiGG IDs that are human-readable shorthand forms of their name (e.g. atp). Components can be explicity referred to using the BiGG ID of the universal component and the charge of the component, separated by a colon (e.g. atp:-4). In practice, the charge can often be omitted in the BiGG ID, since charge is a property of metabolites in SBML models. In the rare case when one would like to use two different components that share the same universal component in a single model, the charge should be explicitly specified in the identifier.
Components are generally expected to represent specific, well-defined molecules. In some cases however, reference compounds describe compounds with a repeating unit (typically polymers). In those cases, the specific value of n (the humber of repeats) is stored in link between the component and reference compound.
When the full formula and/or structure of a component is not known, or not practical to include, components can be defined as a component with a spefic reactive group. This is often the case for reactive groups on proteins or DNA. Since it is often subjective which exact part of the molecule is the reactive group, there is some freedom built into BiGGr for these molecules. When a universal component represents a complex metabolite, the default component has a formula equal to the formula of the reactive part as defined by its reference. Additional components with differnt formulas can be created under the same universal component. BiGG IDs of these components follow the format <universal_id>:<variant><charge>, where the variant is represented by a capital letter (e.g. dnac:A-1).
Collection-specific Metabolites
Collection-specific components and univeral components reside in a separate namespace from the rest of the BiGG IDs, as indicated by their __namespace__ prefix, where namespace is the collection BiGG ID. A component is considered collection-specific when it can not be matched to any of the known metabolites. This causes models that have small errors or peculiarities to not pollute the general namespace. The general BiGG ID namespaces of BiGGr should therefore be of high quality, with clear definitions of each component and reaction.
Compartmentalized Metabolites
A Compartmentalized Component refers to a component within a specific compartment. All metabolites in a model are compartmentalized components. Similarly, a Universal Compartmentalized Component refers to the combination of a universal component and a compartment.
Reactions

A Reaction describes the conversion of compartmentalized components. Similarly to compontents/metabolties, reactions are always a member of a Universal Reaction. Universal reactions group all reactions involving the same pattern of substrates and products, but with optionally more or fewer H+ components at either side of the reaction to balance the charge.
Universal reactions are automatically matched to RHEA reaction or internal references (e.g. for exhange or biomass reactions), based on the reaction pattern and the reference compounds linked to the substrates and products. There is no hard requirement that a reaction should be linked to a reference.

In models, reactions use the BiGG ID of their universal reaction. Whenever a model uses multiple instances of reactions that fall under the same universal reaction, they are numbered using the pattern <BiGG ID>:<copy number>.
Collection-specific Reactions
Analogous to components, reactions can be collection-specific and reside in a separate namespace from the rest of the BiGG IDs, as indicated by their __namespace__ prefix, where namespace is the collection BiGG ID. A reaction is considered collection-specific when it can not be matched to any of the known reactions. This is often the case when one of the components (substrates or products) is collection-specific
Genes & Genomes
Wherever possible, genomes are imported into the database and linked to model genes. No normalization or references are required here.
Features
BiGGr is packed with usefull features, in addition to its core functionality of persenting metabolic models and their components.
Search & Navigation
BiGGr includes a powerfull search function that returns results to the user as fast as possible. Results include any entity where (part of) the BiGG ID, name or synonym matches the search query. BiGGr aims to serve the most relevant results at the top of the results tables. Therefore, BiGG ID matches are preferred over name or synonyms, and exact matches and entities where the start of the BiGG ID, name, etc. matches the search query are prioritized.
In addition, BiGGr search will recognize many types of external identifiers and return the relevant BiGGr entities. For a full list and examples of searching by external identifier, click the search box/button at the top of the page.
MEMOTE Integration

MEMOTE is a suite of tools that can be used to analyze various aspects of model quality. This test suite is now automatically executed for all models in BiGGr. The results can be found on each model page. Whenever MEMOTE results pertain to specific model metabolites or reactions, the results are also displayed on those respective pages.
Escher Maps

Escher is a tool for visualizing metabolic models and pathways. BiGGr uses the normalized BiGG ID namespace to automatically generate model-spefic maps, based on templates or sets of rules.
For maps showing the interconversion of two (or a few) components, all reactions interconverting these components are collected and displayed on the maps. Examples of these maps are Ubiquinone Reduction/Oxidation and L-Arginine Biosynthesis. Other — more complex — maps make use of a template Escher map, on which reactions are mapped (e.g. E. coli Central Metabolism). Maps are required to have a decent completeness (percentage of reactions in the template that are also in the model; exact threshold varies per map), otherwise they are omitted.
License
BiGG is freely available for non-commercial use. It is hosted and maintained by the Systems Biology Research Group at the University of California, San Diego.